Product Description
The QuantSeq-Flex Kit is a library preparation protocol designed to make Illumina compatible libraries from any RNA sample using custom primers. It is open for development by advanced RNA-Seq users based on their custom needs.
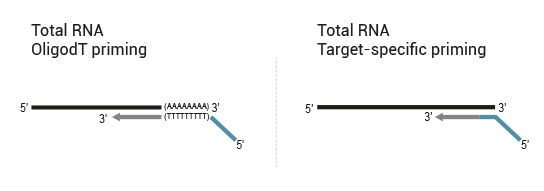
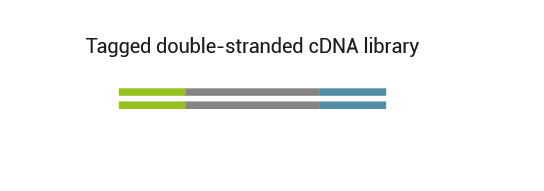
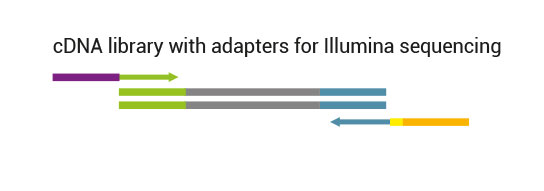
The kit is based on the QuantSeq FWD (Cat. No. 015) strategy, with compatible reagents and Read 1 linker sequence introduced by the Second Strand Synthesis (SSS) primer. Hence the reads generated during the NGS run directly correspond to the RNA sequence.
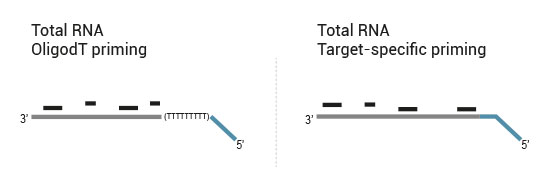
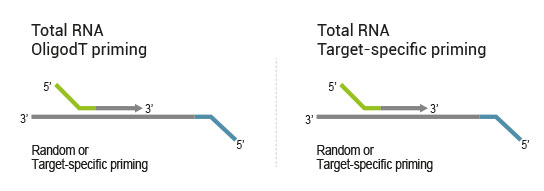
QuantSeq-Flex Kit provides modules for Reverse Transcription (RT) and SSS, allowing users to choose their own primers for either one of these steps or both of them. This offers maximum flexibility in the choice of targets and input material.
With this highly flexible QuantSeq kit the following types of libraries can be generated:
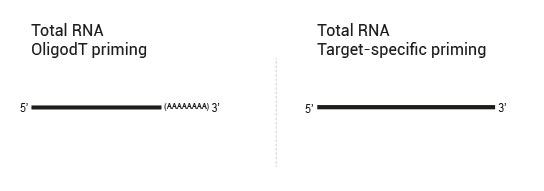
- 1. OligodT primed during RT, random primed during SSS (QuantSeq 3’ mRNA-Seq)
- 2. OligodT primed during RT, target-specifically primed during SSS (targeted 3’ mRNA-Seq)
- 3. Target-specifically primed during RT, random primed during SSS (targeted RNA-Seq, allows for identification of novel fusions)
- 4. Target-specifically primed during RT, target-specifically primed during SSS (targeted RNA-Seq, known targets detectable only)
Whether it is gene expression analysis, targeted sequencing, adapter ligation-based RNA-Seq or any other experiment where a certain RNA region needs to be sequenced, QuantSeq-Flex can be used together with user-supplied primers.
Multiplexing
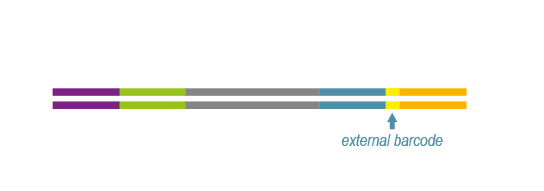
QuantSeq libraries are intended for a high degree of multiplexing. External barcodes allowing up to 96 samples to be sequenced per lane on an Illumina flow cell are included in the kit. This high level of multiplexing allows saving costs as the length restriction in QuantSeq saves sequencing space.
In order to be compatible with the supplied barcode primers please note our recommendations for the custom primer design (Refer User Guide: Appendix B- Primer design)
For the detailed information about barcodes and instructions how to use them please refer User Guide: Appendix F- Multiplexing, QuantSeq-Flex User Guide).
An RNA-Seq experiment tailored to your needs
QuantSeq-Flex modular kit gives maximum flexibility to the user encouraging implementation of RNA-Seq as a part of the experimental pipeline. One fragment per transcript produced and usage of custom primers makes this kit a perfect solution for cost-saving high-throughput gene expression analysis. QuantSeq-Flex is suitable for targeted sequencing of gene panels, enrichment, detection of fusion genes and various other types of transcripts.