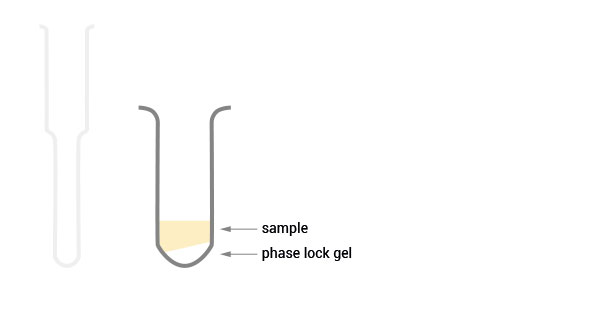
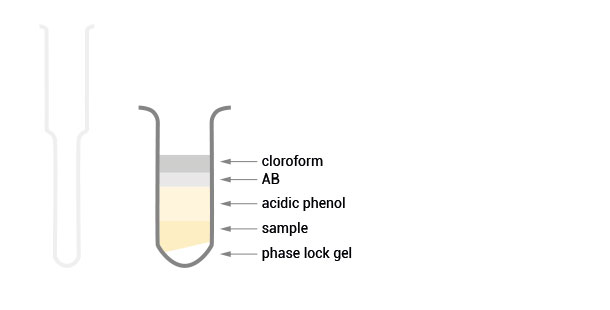
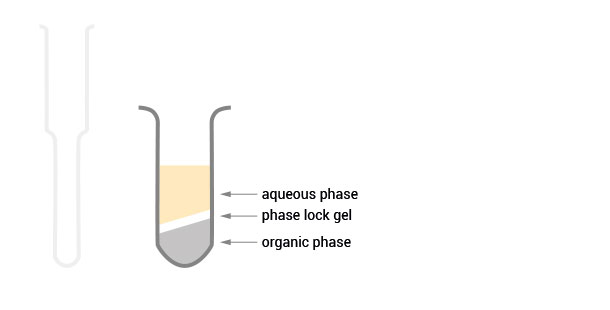
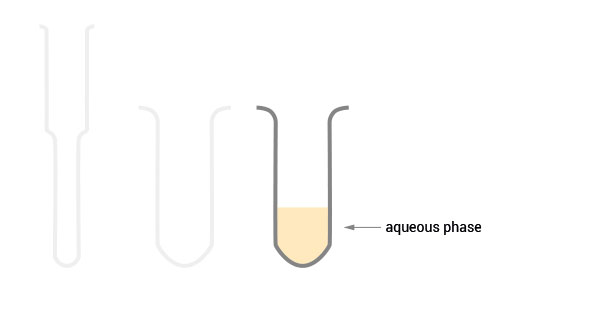
Product Description
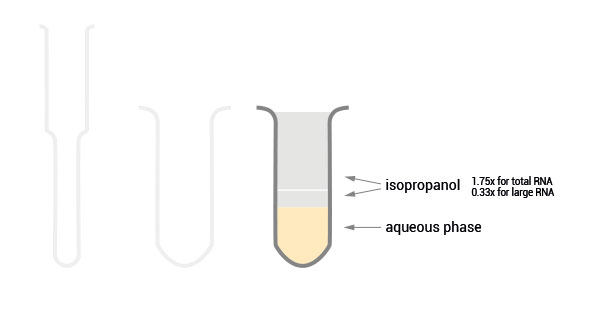
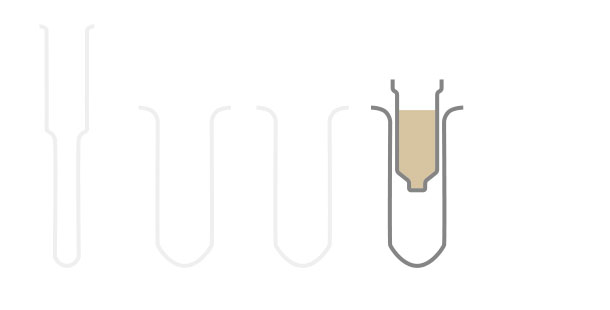
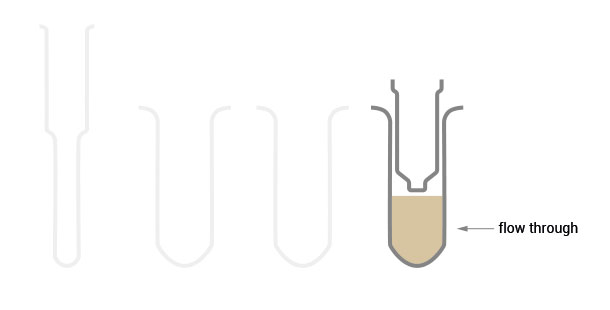
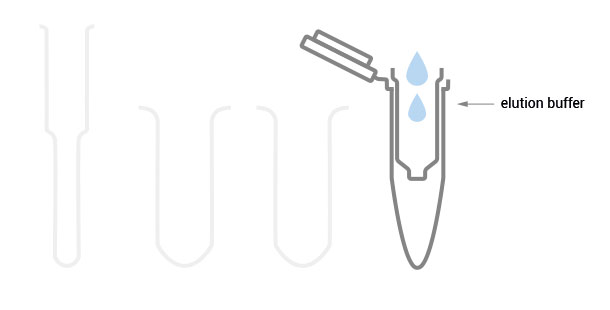
The SPLIT RNA Extraction Kit enables a fast and highly efficient extraction of RNA that is free of genomic DNA contamination. The RNA can be recovered as total RNA or split into two fractions, large RNA and small RNA, facilitating the analysis of e.g., mRNA and miRNA from the same sample. Thus the RNA obtained is ideal for seamlessly preparing libraries for Next Generation Sequencing of total RNA or its large and small fractions or any other demanding downstream application.